INCISIVE Implementation Guide
0.1.1 - draft
INCISIVE Implementation Guide
0.1.1 - draft
INCISIVE Implementation Guide - Local Development build (v0.1.1) built by the FHIR (HL7® FHIR® Standard) Build Tools. See the Directory of published versions
| Official URL: https://fhir.incisive-project.eu/StructureDefinition/ProstateTumorProfileResultLogicalModel | Version: 0.1.1 | |||
| Draft as of 2024-03-19 | Computable Name: ProstateTumorProfileResultLogicalModel | |||
INCISIVE based logical model for the tumor profile result of Prostate cancer
Usage:
Description of Profiles, Differentials, Snapshots and how the different presentations work.
This structure is derived from Base
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | Base | Tumor profile result Prostate Instances of this logical model are not marked to be the target of a Reference | |
  | 1..1 | Identifier | An identifier for the tumor profile result | |
  | 1..1 | INCISIVEPatientLogicalModel | INCISIVE patient | |
  | 0..1 | ProstateHistologyMutationsLogicalModel | Histology - Mutations reference | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | boolean | Cribriform neoplasm pattern (finding) | |
  | 0..1 | boolean | Non-infiltrating intraductal carcinoma (morphologic abnormality) | |
  | 0..1 | decimal | % of affected cores in right lobe | |
  | 1..1 | decimal | % of affected cores in the left lobe | |
  | 0..1 | integer | Histologic grade of urothelial carcinoma by World Health Organization and International Society of Urological Pathology technique (observable entity) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3 (qualifier value) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3b (qualifier value) | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological T category allowable value (qualifier value) | |
  | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological N category allowable value (qualifier value) | |
 Documentation for this format Documentation for this format | ||||
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | Base | Tumor profile result Prostate Instances of this logical model are not marked to be the target of a Reference | |
  | 1..1 | Identifier | An identifier for the tumor profile result | |
  | 1..1 | INCISIVEPatientLogicalModel | INCISIVE patient | |
  | 0..1 | ProstateHistologyMutationsLogicalModel | Histology - Mutations reference | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | boolean | Cribriform neoplasm pattern (finding) | |
  | 0..1 | boolean | Non-infiltrating intraductal carcinoma (morphologic abnormality) | |
  | 0..1 | decimal | % of affected cores in right lobe | |
  | 1..1 | decimal | % of affected cores in the left lobe | |
  | 0..1 | integer | Histologic grade of urothelial carcinoma by World Health Organization and International Society of Urological Pathology technique (observable entity) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3 (qualifier value) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3b (qualifier value) | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological T category allowable value (qualifier value) | |
  | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological N category allowable value (qualifier value) | |
 Documentation for this format Documentation for this format | ||||
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | Base | Tumor profile result Prostate Instances of this logical model are not marked to be the target of a Reference | |
  | 1..1 | Identifier | An identifier for the tumor profile result | |
  | 1..1 | INCISIVEPatientLogicalModel | INCISIVE patient | |
  | 0..1 | ProstateHistologyMutationsLogicalModel | Histology - Mutations reference | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | boolean | Cribriform neoplasm pattern (finding) | |
  | 0..1 | boolean | Non-infiltrating intraductal carcinoma (morphologic abnormality) | |
  | 0..1 | decimal | % of affected cores in right lobe | |
  | 1..1 | decimal | % of affected cores in the left lobe | |
  | 0..1 | integer | Histologic grade of urothelial carcinoma by World Health Organization and International Society of Urological Pathology technique (observable entity) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3 (qualifier value) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3b (qualifier value) | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological T category allowable value (qualifier value) | |
  | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological N category allowable value (qualifier value) | |
 Documentation for this format Documentation for this format | ||||
This structure is derived from Base
Differential View
This structure is derived from Base
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | Base | Tumor profile result Prostate Instances of this logical model are not marked to be the target of a Reference | |
  | 1..1 | Identifier | An identifier for the tumor profile result | |
  | 1..1 | INCISIVEPatientLogicalModel | INCISIVE patient | |
  | 0..1 | ProstateHistologyMutationsLogicalModel | Histology - Mutations reference | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | boolean | Cribriform neoplasm pattern (finding) | |
  | 0..1 | boolean | Non-infiltrating intraductal carcinoma (morphologic abnormality) | |
  | 0..1 | decimal | % of affected cores in right lobe | |
  | 1..1 | decimal | % of affected cores in the left lobe | |
  | 0..1 | integer | Histologic grade of urothelial carcinoma by World Health Organization and International Society of Urological Pathology technique (observable entity) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3 (qualifier value) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3b (qualifier value) | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological T category allowable value (qualifier value) | |
  | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological N category allowable value (qualifier value) | |
 Documentation for this format Documentation for this format | ||||
Key Elements View
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | Base | Tumor profile result Prostate Instances of this logical model are not marked to be the target of a Reference | |
  | 1..1 | Identifier | An identifier for the tumor profile result | |
  | 1..1 | INCISIVEPatientLogicalModel | INCISIVE patient | |
  | 0..1 | ProstateHistologyMutationsLogicalModel | Histology - Mutations reference | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | boolean | Cribriform neoplasm pattern (finding) | |
  | 0..1 | boolean | Non-infiltrating intraductal carcinoma (morphologic abnormality) | |
  | 0..1 | decimal | % of affected cores in right lobe | |
  | 1..1 | decimal | % of affected cores in the left lobe | |
  | 0..1 | integer | Histologic grade of urothelial carcinoma by World Health Organization and International Society of Urological Pathology technique (observable entity) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3 (qualifier value) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3b (qualifier value) | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological T category allowable value (qualifier value) | |
 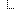 | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological N category allowable value (qualifier value) | |
 Documentation for this format Documentation for this format | ||||
Snapshot View
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | Base | Tumor profile result Prostate Instances of this logical model are not marked to be the target of a Reference | |
  | 1..1 | Identifier | An identifier for the tumor profile result | |
  | 1..1 | INCISIVEPatientLogicalModel | INCISIVE patient | |
  | 0..1 | ProstateHistologyMutationsLogicalModel | Histology - Mutations reference | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | boolean | Cribriform neoplasm pattern (finding) | |
  | 0..1 | boolean | Non-infiltrating intraductal carcinoma (morphologic abnormality) | |
  | 0..1 | decimal | % of affected cores in right lobe | |
  | 1..1 | decimal | % of affected cores in the left lobe | |
  | 0..1 | integer | Histologic grade of urothelial carcinoma by World Health Organization and International Society of Urological Pathology technique (observable entity) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3 (qualifier value) | |
  | 0..1 | boolean | American Joint Committee on Cancer pT3b (qualifier value) | |
  | 0..1 | integer | Gleason score (observable entity) | |
  | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological T category allowable value (qualifier value) | |
 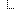 | 0..1 | CodeableConcept | American Joint Committee on Cancer pathological N category allowable value (qualifier value) | |
 Documentation for this format Documentation for this format | ||||
This structure is derived from Base